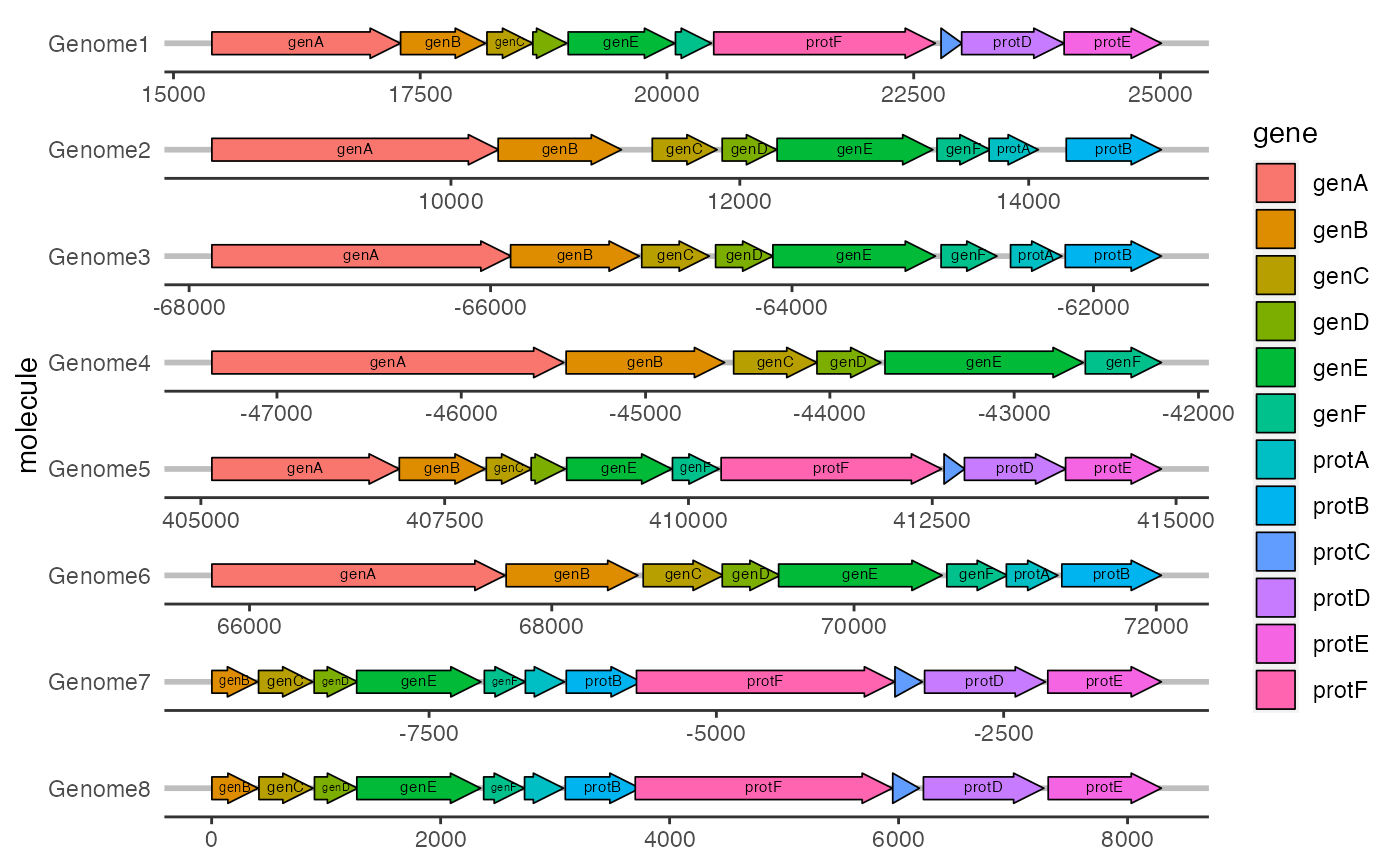
geom_gene_label() can be used to add a text label to genes drawn with
geom_gene_arrow().
geom_gene_label(
mapping = NULL,
data = NULL,
stat = "identity",
position = "identity",
na.rm = FALSE,
show.legend = FALSE,
inherit.aes = TRUE,
padding.x = grid::unit(1, "mm"),
padding.y = grid::unit(0.1, "lines"),
align = "centre",
min.size = 4,
grow = F,
reflow = F,
height = grid::unit(3, "mm"),
...
)Arguments
- mapping, data, stat, position, na.rm, show.legend, inherit.aes, ...
Standard geom arguments as for
ggplot2::geom_text().- padding.x, padding.y
grid::unit()object, giving horizontal or vertical padding around the text. Defaults to 1 mm and 0.1 lines respectively.- align
Where inside the gene to place the text label. Default is 'centre'; other options are 'left' and 'right'.
- min.size
Minimum font size, in points. If provided, text that would need to be shrunk below this size to fit inside the gene arrow will not be drawn. Defaults to 4 pt.
- grow
If
TRUE, text will be grown as well as shrunk to fill the arrow.- reflow
If
TRUE, text will be reflowed (wrapped) to better fit the arrow.- height
grid::unit()object giving the maximum height of the text. Defaults to 3 mm, which is the default height of gene arrows drawn withgeom_gene_arrow().
Details
geom_gene_label() uses the 'ggfittext' package to fit text to genes. All
text drawing options available in ggfittext::geom_fit_text() (growing,
reflowing, etc.) are also available here. For full details on how these
options work, see the documentation for ggfittext::geom_fit_text().
Standard 'ggplot2' aesthetics for text are supported (see Aesthetics).
Aesthetics
xmin,xmax (start and end of the gene; required)
y (molecule; required)
label (the label text; required)
colour
size
alpha
family
fontface
angle
See also
geom_gene_arrow
Examples
ggplot2::ggplot(example_genes, ggplot2::aes(xmin = start, xmax = end,
y = molecule, fill = gene, label = gene)) +
geom_gene_arrow() +
geom_gene_label() +
ggplot2::facet_wrap(~ molecule, ncol = 1, scales = "free") +
theme_genes()